The nucleotides (As, Ts, Cs, and Gs) of deoxyribonucleic acid (DNA) are copied in a process called DNA replication, which is performed by a complex of molecular machinery called the replisome. The coding strand of double stranded DNA acts as a blueprint for the replisome, storing information that is critical for explaining how to create sub-cellular molecules like proteins. This information-storage feature of the DNA coding strand is precious, because one small change in the sequence of nucleotides making up the DNA coding strand could result in a mutation that is harmful to the cell or organism.
To guard against mutations, all cells contain various DNA repair systems. DNA repair works to preserve the information in the DNA coding strand by maintaining the order of nucleotides. For decades, a protein known as MutS has been shown to play a key role in DNA repair. MutS binds to mispaired bases or “errors” that occur in the DNA sequence and initiates their correction. Like searching for a needle in a haystack, MutS must search for and identify a single error in the DNA sequence that occurs just once in tens of millions of correct pairings.
For many years, the process by which MutS searches for an error in the DNA sequence has been unknown to scientists, because they have lacked the resolution necessary to detect the searching behavior of MutS. A research collaboration between University of Michigan Associate Professor Dr. Lyle Simmons, Assistant Professor Dr. Julie Biteen, and their co-workers in the Departments of Molecular, Cellular, and Developmental Biology and Chemistry, resulted in a recent publication in the Proceedings of the National Academy of Sciences (PNAS). It details the discovery of how a single MutS molecule conducts its search for DNA in need of repair inside a living bacterial cell.
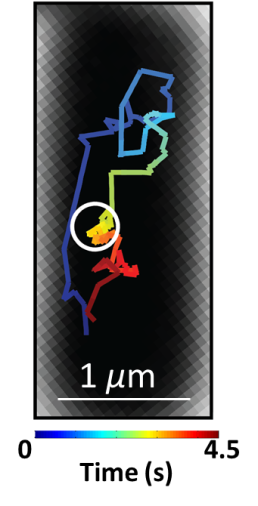 Using real time super-resolution microscopy and single-molecule tracking in living bacterial cells, the Simmons and Biteen research teams discovered MutS moves around the cell very rapidly looking for DNA in need of repair. Yet, as seen in this image on the left depicting the movement of MutS in a bacterial cell (shown in black), MutS also moves to the replisome (circled in white) and pauses to search for errors before moving away. The behavior of MutS is indicated in the image by tracking its movements (colored purple to red). Strikingly, the Simmons and Biteen research teams show that MutS is also positioned at the replisome in cells during normal growth, prior to the formation of an error in DNA. Such a localized search behavior at the replisome by MutS allows for the rapid and efficient recognition of errors in the DNA sequence, and allows MutS to initiate DNA repair before those errors become permanent mutations. Dr. Simmons notes, “This work provides a new fundamental insight into how MutS searches for rare base-pairing errors in the complex environment of a living cell.”
Using real time super-resolution microscopy and single-molecule tracking in living bacterial cells, the Simmons and Biteen research teams discovered MutS moves around the cell very rapidly looking for DNA in need of repair. Yet, as seen in this image on the left depicting the movement of MutS in a bacterial cell (shown in black), MutS also moves to the replisome (circled in white) and pauses to search for errors before moving away. The behavior of MutS is indicated in the image by tracking its movements (colored purple to red). Strikingly, the Simmons and Biteen research teams show that MutS is also positioned at the replisome in cells during normal growth, prior to the formation of an error in DNA. Such a localized search behavior at the replisome by MutS allows for the rapid and efficient recognition of errors in the DNA sequence, and allows MutS to initiate DNA repair before those errors become permanent mutations. Dr. Simmons notes, “This work provides a new fundamental insight into how MutS searches for rare base-pairing errors in the complex environment of a living cell.”
Dr. Simmons and his lab members bring their excitement about science discovery from their laboratory to elementary, high school, and university classes through hands on experiments in K-5 classes, analysis of genomic data with high school students, and design of new curriculum to ensure the success of diverse students pursuing science PhDs.
This work is funded by the Division of Molecular and Cellular Biosciences, Award #MCB-1050948.
